Help
Browse Omnicrobe
All on-line services are available in the navbar menu and on the Omnicrobe main page. The results of the user queries are displayed as tables as below.
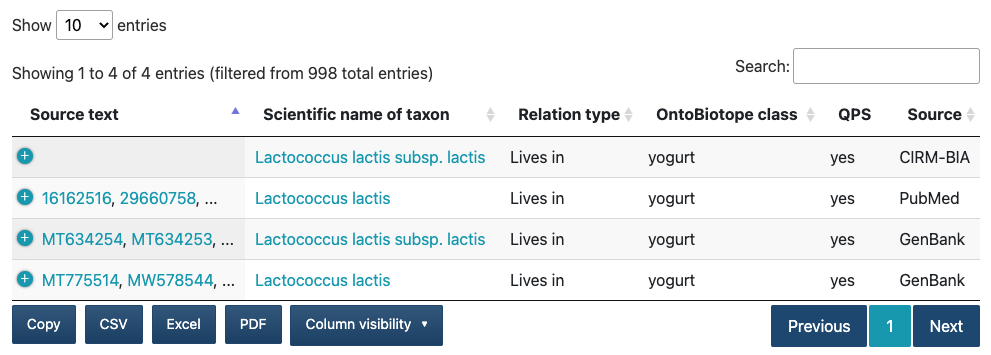
The Source text column lists the first two identifiers of the documents where the information is extracted from. A click on a document identifier opens the document as made available by the source provider. A click on the icon of the beginning column displays the whole list of document identifiers in the Full source text column.
The Scientific name of taxon column displays the scientific name of the taxon. A click on the taxon name displays the NCBI taxonomy browser or the DSMZ BacDive description.
The OntoBiotope class column displays the OntoBiotope class name of the habitat, phenotype or use.
The QPS column displays the QPS (Qualified Presumption of Safety) EFSA label. If the value is yes, the microorganism involved in the relation has a QPS EFSA label.
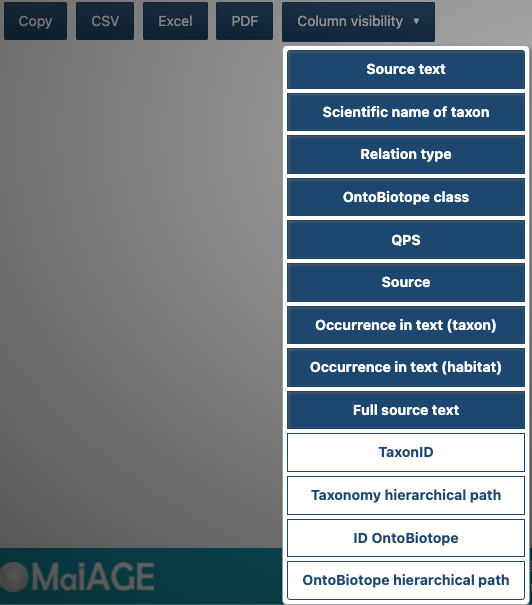
The Column visibility button allows you to select other information of interest to be displayed in the columns of the table such as the TaxonID (taxonomy identifier), the Taxonomy hierarchical path, the ID OntoBiotope (OntoBiotope identifier) and the OntoBiotope hierarchical path.
For the API, see API documentation.
About habitats
Display the habitats for a microorganism
Select Taxon lives in Habitat in the Search menu.
Fill the search box Search relations by taxon with the microorganism name. It can be any taxon (e.g. genus, family, including strain).
The search box suggests completions for the name. Select the relevant taxon.
The list of results includes Document and Taxon (see Browse Omnicrobe), and Habitat.
The OntoBiotope class column displays the OntoBiotope class name of the habitat. A click on the first column displays the whole list of forms it has in the source text in the Occurrence in text (habitat) column.
Filter given types of habitats among microorganism habitats
Select Taxon lives in Habitat in the Search menu.
Fill the search box Search relations by taxon with the microorganism name. It can be any taxon (e.g. genus, family, including strain).
The search box suggests completions for the name. Select the relevant taxon.
Click on The Filter selection button. It displays additional criteria to restrict the results.
Fill the search box Filter by sources with one or more sources separated by ";".
Fill the search box Filter by habitat with one or more habitat names separated by ";".
Check the QPS checkbox to filter only microorganisms with a QPS EFSA label.
Display the microorganism living in an habitat
Select Habitat contains Taxon in the Search menu.
Fill the search box Search relations by habitat with the habitat name. It can be a general or a specific habitat (e.g. 'food', 'cheese brine').
The search box suggests completions for the name. Select the relevant habitat.
The list of results includes Document and Taxon (see Browse Omnicrobe), and Habitat.
The OntoBiotope class column displays the OntoBiotope class name of the habitat. A click on the first column displays the whole list of forms it has in the source text in the Occurrence in text (habitat) column.
Filter given microorganisms among the microorganisms of an habitat
Select Habitat contains Taxon in the Search menu.
Fill the search box Search relations by habitat with the habitat name. It can be a general or a specific habitat (e.g. 'food', 'cheese brine').
The search box suggests completions for the name. Select the relevant habitat.
Click on The Filter selection button. It displays additional criteria to restrict the results.
Fill the search box Filter by sources with one or more sources separated by ";".
Fill the search box Filter by taxon with one or more taxon names separated by ";".
Check the QPS checkbox to filter only microorganisms with a QPS EFSA label.
I do not know how to name the habitat that I search for
Select Habitat contains Taxon in the Search menu.
Click on the OntoBiotope navigation link and search the habitat name in the search class or click on triangle-shaped icons to browse the OntoBiotope ontology. It opens the habitat classification.
Click on the habitat name to copy it in the search box and send the query.
About Phenotypes
Display the phenotypes of a microorganism
Select Taxon exhibits Phenotype in the Search menu.
Fill the search box Search relations by taxon with the microorganism name. It can be any taxon (e.g. genus, family, including strain).
The search box suggests completions for the name. Select the relevant taxon.
The list of results includes Document and Taxon (see Browse Omnicrobe), and Phenotype.
The OntoBiotope class column displays the OntoBiotope class name of the phenotype. A click on the first column displays the whole list of forms it has in the source text in the Occurrence in text (phenotype) column.
Filter given types of phenotypes among microorganism phenotypes
Select Taxon exhibits Phenotype in the Search menu.
Fill the search box Search relations by taxon with the microorganism name. It can be any taxon (e.g. genus, family, including strain).
The search box suggests completions for the name. Select the relevant taxon.
Click on The Filter selection button. It displays additional criteria to restrict the results.
Fill the search box Filter by sources with one or more sources separated by ";".
Fill the search box Filter by phenotype with one or more phenotype names separated by ";".
Check the QPS checkbox to filter only microorganisms with a QPS EFSA label.
Display the microorganisms that express a given phenotype
Select Phenotype is exhibited by Taxon in the Search menu.
Fill the search box Search relations by phenotype with the phenotype name. It can be a general or a specific phenotype (e.g. 'physiological phenotype', 'obligate piezophile').
The search box suggests completions for the name. Select the relevant phenotype.
The list of results includes Document and Taxon (see Browse Omnicrobe), and Phenotype.
The OntoBiotope class column displays the OntoBiotope class name of the phenotype. A click on the first column displays the whole list of forms it has in the source text in the Occurrence in text (phenotype) column.
Filter given microorganisms among the microorganisms that express a given phenotype
Select Phenotype is exhibited by Taxon in the Search menu.
Fill the search box Search relations by phenotype with the phenotype name. It can be a general or a specific phenotype (e.g. 'physiological phenotype', 'obligate piezophile').
The search box suggests completions for the name. Select the relevant phenotype.
Click on The Filter selection button. It displays additional criteria to restrict the results.
Fill the search box Filter by sources with one or more sources separated by ";".
Fill the search box Filter by taxon with one or more taxon names separated by ";".
Check the QPS checkbox to filter only microorganisms with a QPS EFSA label.
I do not know how to name the phenotype that I search for
Select Phenotype is exhibited by Taxon in the Search menu.
Click on the OntoBiotope navigation link and search the phenotype name in the search class or click on triangle-shaped icons to browse the OntoBiotope ontology. It opens the phenotype classification.
Click on the phenotype name to copy it in the search box and send the query.
About uses
Display the uses of a microorganism
Select Taxon studied for use in the Search menu.
Fill the search box Search relations by taxon with the microorganism name. It can be any taxon (e.g. genus, family, including strain).
The search box suggests completions for the name. Select the relevant taxon.
The list of results includes Document and Taxon (see Browse Omnicrobe), and Use.
The OntoBiotope class column displays the OntoBiotope class name of the use. A click on the first column displays the whole list of forms it has in the source text in the source text in the Occurrence in text (use).
Filter given types of uses among microorganism uses
Select Taxon studied for use in the Search menu.
Fill the search box Search relations by taxon with the microorganism name. It can be any taxon (e.g. genus, family, including strain).
The search box suggests completions for the name. Select the relevant taxon.
Click on The Filter selection button. It displays additional criteria to restrict the results.
Fill the search box Filter by sources with one or more sources separated by ";".
Fill the search box Filter by use with one or more use names separated by ";".
Check the QPS checkbox to filter only microorganisms with a QPS EFSA label.
Display the microorganism given a use
Select Use involves Taxon in the Search menu.
Fill the search box Search relations by use with the use name. It can be a general or a specific use (e.g. 'sensory quality', 'viscosity').
The search box suggests completions for the name. Select the relevant use.
The list of results includes Document and Taxon (see Browse Omnicrobe), and Use.
The OntoBiotope class column displays the OntoBiotope class name of the use. A click on the last column displays the whole list of forms it has in the source text in the source text in the Occurrence in text (use).
Filter microorganisms among the microorganisms that are studied for a given use
Select Use involves Taxon in the Search menu.
Fill the search box Search relations by use with the use name. It can be a general or a specific use (e.g. 'sensory quality', 'viscosity').
The search box suggests completions for the name. Select the relevant use.
Click on The Filter selection button. It displays additional criteria to restrict the results.
Fill the search box Filter by sources with one or more sources separated by ";".
Fill the search box Filter by taxon with one or more taxon names separated by ";".
Check the QPS checkbox to filter only microorganisms with a QPS EFSA label.
I do not know how to name the use that I search for
Select Use involves Taxon in the Search menu.
Click on the OntoBiotope navigation link and search the use name in the search class or click on triangle-shaped icons to browse the OntoBiotope ontology. It opens the use classification.
Click on the use name to copy it in the search box and send the query.
Advanced functionalities
Export the results
Below the table, click on CSV or Excel button.
Select the columns to be exported by clicking on the Column visibility button (unselected columns are white).
Using the API is recommended for the export of large set of data.
Advanced search
Select Advanced search in the Search menu.
Select from the list the type of element to be searched (taxon, habitat, phenotype, use) and fill the search box with the name of the element.
Click on Add rule to add one or more items to the query.
Click on Add group to build groups of items in the query.
Click on AND or OR to select the operator between items in the query.
Click on Apply button to send a query.
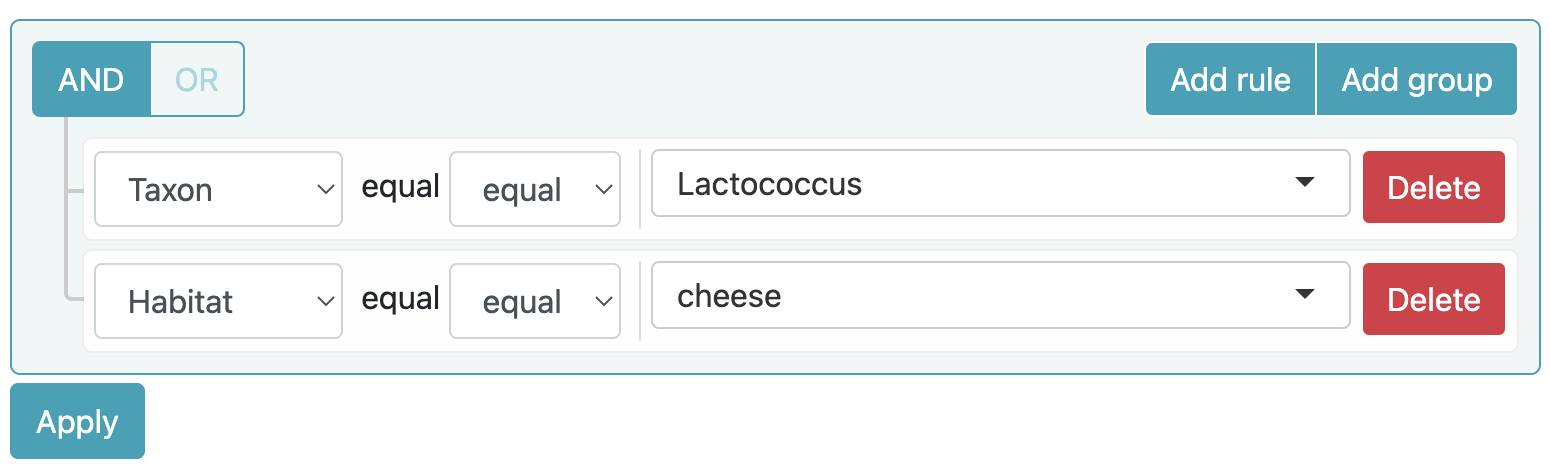
Example 1
Search all relations involving the lactococcus taxon and the cheese habitat.
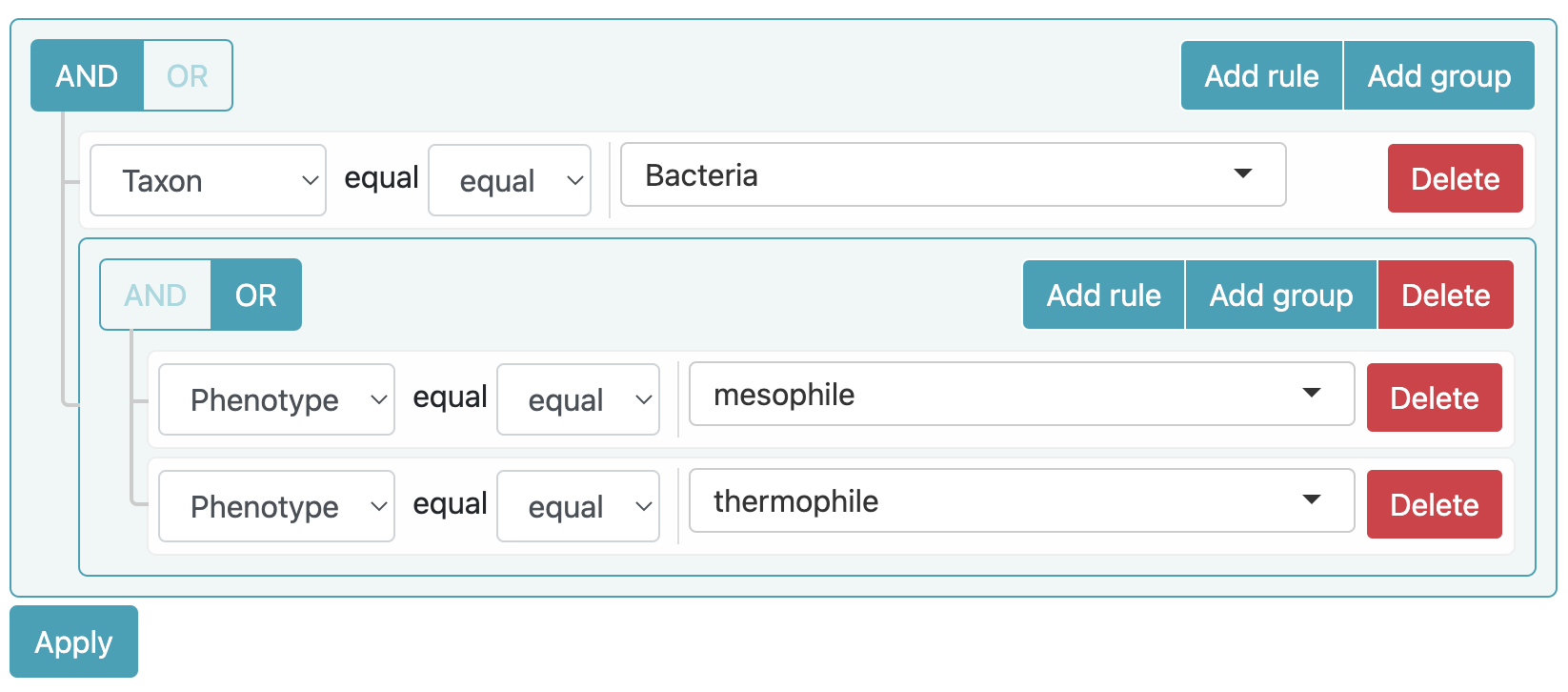
Example 2
Search all relations involving the mesophile phenotype or the thermophile phenotype, and the Bacteria taxon.
Learn more
On Omnicrobe project
Click on the About menu to learn on the text mining process, the Ontobiotope classification, the partners and the publications.
On Omnicrobe data
The sources are scientific papers, catalogue of biological resource centers and DNA sequence database.
Click on the About Omnicrobe tab. The Current version section describes the versions of the data and the volumes.
The Public data section describes the sources of the data. The Sources section of Terms of use and Copyright describes the licenses for each source.
Providing a feedback
The results are automatically extracted and interpreted by machine learning algorithms of the Alvis platform. Its quality is continuously improving but some errors may remain.
To request their correction: omnicrobe@inrae.fr.

